

- #GENEIOUS PRIME 2020 INSTALL#
- #GENEIOUS PRIME 2020 PASSWORD#
Fixed occasionally being unable to uninstall Geneious. Fixed a bug where ctrl+drag (or alt+drag) documents into the same folder and choosing 'Replace' deletes these documents. Added ability for admins to disable arbitrary plugins. #GENEIOUS PRIME 2020 PASSWORD#
Added a button to hide/show text on all password fields.Geneious Server: Now ensures python3 is available for SPAdes.Improve performance when copying multiple documents and folders in the local database.
#GENEIOUS PRIME 2020 INSTALL#
Prevented attempts to install an incompatible version of python.Fixed annotation colors not showing up sometimes when Find CRISPR Sites is run on sequence lists.Better detection of pre-installed python3.Added parameter -installationId to display installation id in command line.Sequence Lists produce correct lineage interval in Golden Gate cloning.Sequence Lists are now reversed correctly in Golden Gate cloning.Operations should now perform significantly faster on large datasets.Fixed rare crash where rearranging cloning inserts in restriction cloning.Fixed a bug where cloning results had the wrong insert order in their names.Chromatogram Viewer: Fixed a bug where some Color Schemes were showing incorrect line and text colors.Fixed bug where changing options in BLAST workflows for one blast type affected other blast type options.Apply Variants to Reference Sequence: Added option to mask low coverage regions with N.Advanced Preferences: Fixed advanced preference table values not being reloaded after changes.Added floating name column to document and annotation tables.Added filter functionality to dropdown menus.Improved the naming scheme for sequence list results of cloning operations.Improved input validation for batch operations.3D Protein viewer: Updated Jmol to version 6.Minor Features and Usability Improvements:
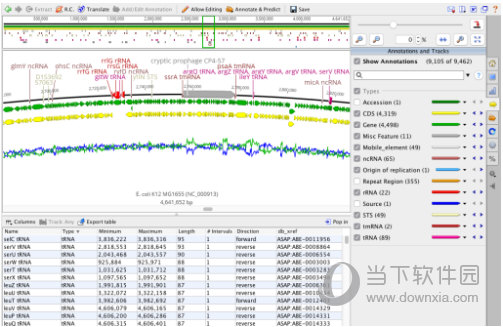 Primer Import: Added the ability to automatically annotate primer extensions on import if provided appropriately formatted sequences. Parts Cloning: Added support for protein documents. Command Line Interface: Added support to import and export files from shared databases. Command Line Interface: Added support to import and export files from local databases. Our results demonstrate that SARS-CoV-2-neutralizing antibodies are readily generated from a diverse pool of precursors, fostering hope for rapid induction of a protective immune response upon vaccination.Geneious Prime version history and release notes Release notes for Geneious Primeġ0.0 10.1 10.2 11.0 11.1 2019.0 2019.1 2019.2 2020.0 2020.1 2020.2 2021.0 2021.1 2021.2 2022.0 2022.1 2022.2 Version 2022.2 Beta (14 June 2022) Interestingly, potential precursor sequences were identified in naive B cell repertoires from 48 healthy individuals who were sampled before the COVID-19 pandemic. Of these, 27 potently neutralized authentic SARS-CoV-2 with IC 100 as low as 0.04 μg/mL, showing a broad spectrum of variable (V) genes and low levels of somatic mutations. By screening 4,313 SARS-CoV-2-reactive B cells, we isolated 255 antibodies from different time points as early as 8 days after diagnosis. To identify SARS-CoV-2-neutralizing antibodies, we analyzed the antibody response of 12 COVID-19 patients from 8 to 69 days after diagnosis. Because approved drugs and vaccines are limited or not available, new options for COVID-19 treatment and prevention are in high demand. The SARS-CoV-2 pandemic has unprecedented implications for public health, social life, and the world economy.
Primer Import: Added the ability to automatically annotate primer extensions on import if provided appropriately formatted sequences. Parts Cloning: Added support for protein documents. Command Line Interface: Added support to import and export files from shared databases. Command Line Interface: Added support to import and export files from local databases. Our results demonstrate that SARS-CoV-2-neutralizing antibodies are readily generated from a diverse pool of precursors, fostering hope for rapid induction of a protective immune response upon vaccination.Geneious Prime version history and release notes Release notes for Geneious Primeġ0.0 10.1 10.2 11.0 11.1 2019.0 2019.1 2019.2 2020.0 2020.1 2020.2 2021.0 2021.1 2021.2 2022.0 2022.1 2022.2 Version 2022.2 Beta (14 June 2022) Interestingly, potential precursor sequences were identified in naive B cell repertoires from 48 healthy individuals who were sampled before the COVID-19 pandemic. Of these, 27 potently neutralized authentic SARS-CoV-2 with IC 100 as low as 0.04 μg/mL, showing a broad spectrum of variable (V) genes and low levels of somatic mutations. By screening 4,313 SARS-CoV-2-reactive B cells, we isolated 255 antibodies from different time points as early as 8 days after diagnosis. To identify SARS-CoV-2-neutralizing antibodies, we analyzed the antibody response of 12 COVID-19 patients from 8 to 69 days after diagnosis. Because approved drugs and vaccines are limited or not available, new options for COVID-19 treatment and prevention are in high demand. The SARS-CoV-2 pandemic has unprecedented implications for public health, social life, and the world economy.






 0 kommentar(er)
0 kommentar(er)
